
To close the gene synthesis gap, we’ve returned to first principles.

Our technology puts accurate gene synthesis capabilities directly into the hands of researchers. To do this, we’ve developed a method for synthesizing, assembling and error-correcting gene-length DNA on a single silicon chip within a benchtop device.
Our technology will enable scale and flexibility far beyond the limits of centralized service labs. DNA availability will no longer be a bottleneck. Instead, it’ll provide opportunities to design more ambitious experiments and deliver faster results.
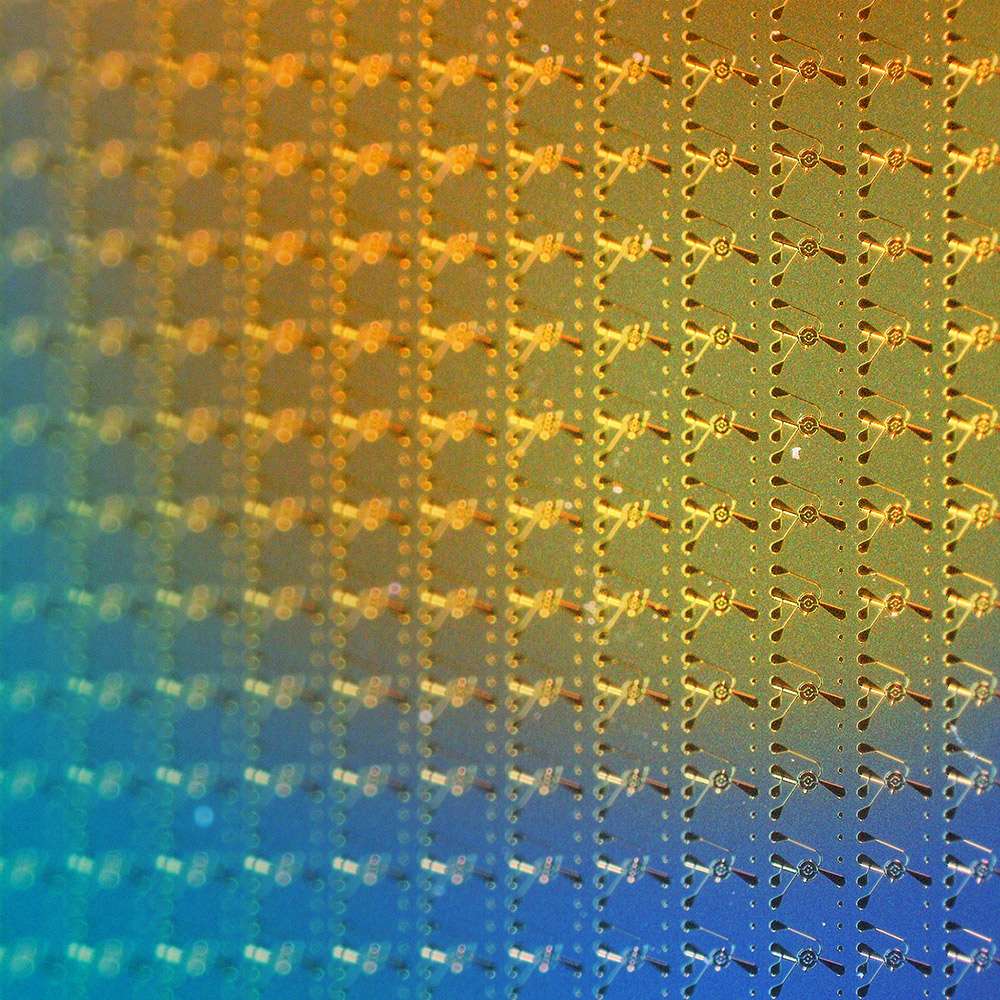
Thermal control of DNA synthesis
We have re-engineered phosphoramidite chemistry to enhance the precision of DNA synthesis through thermal control of oligonucleotide extension.
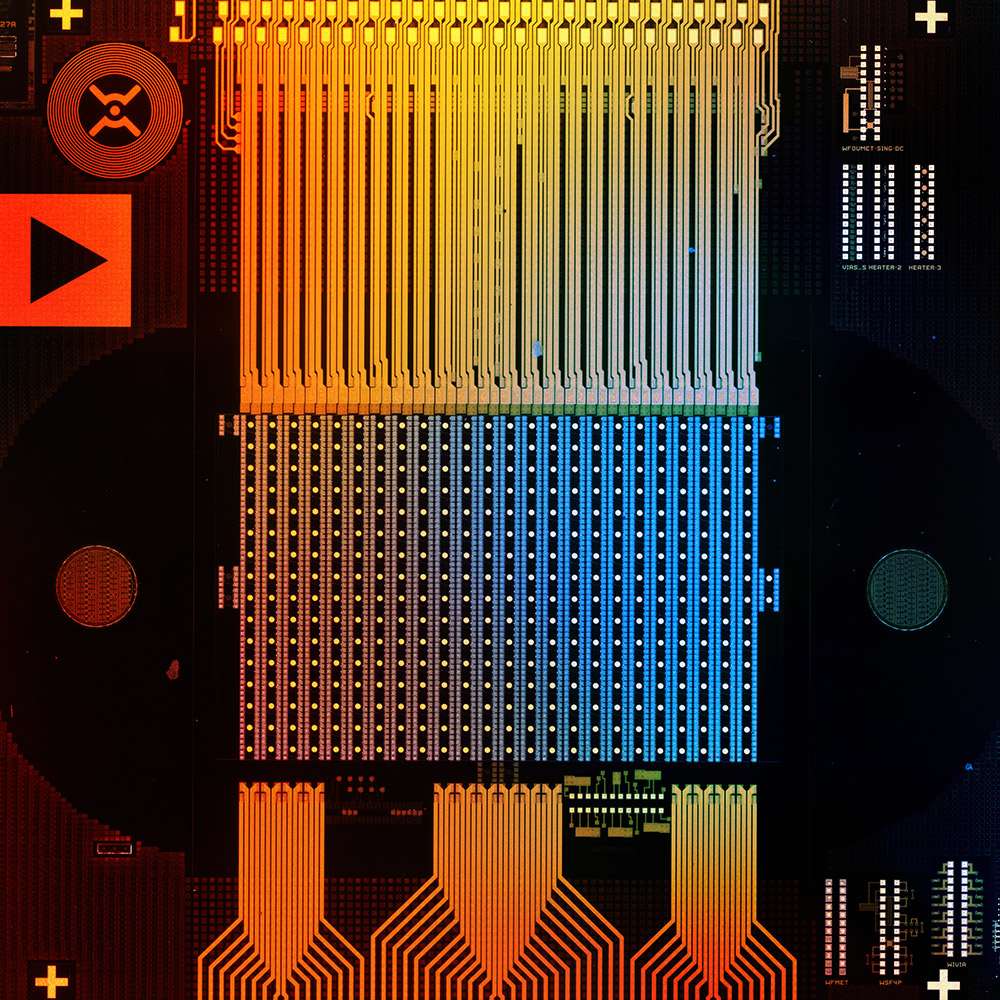
Novel chip technology for highly parallel DNA synthesis
We have designed unique DNA synthesis chips capable of synthesizing thousands of independent oligonucleotide sequences simultaneously under thermal control.
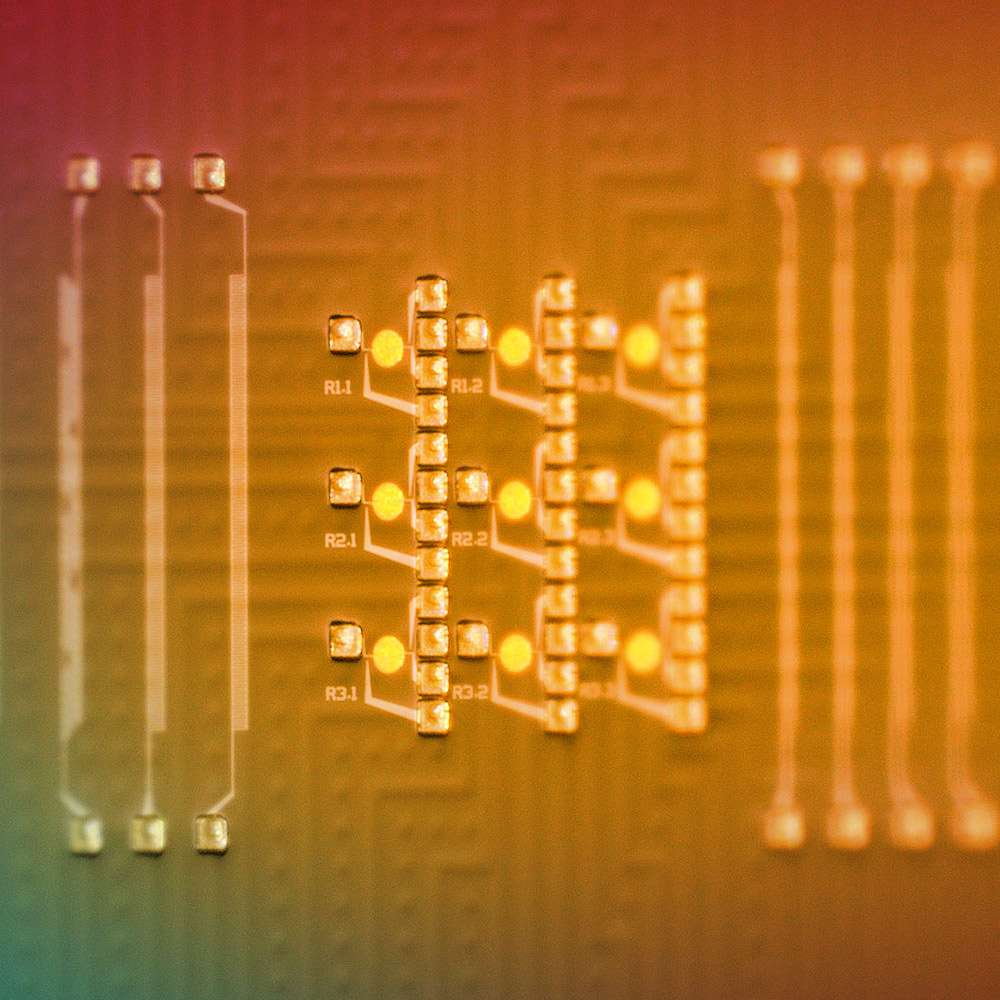
Our Binary Assembly® process generates long accurate DNA in just hours
We assemble individual oligonucleotides into double-stranded gene-length sequences at the surface of our chip through the ordered, sequential pairing of complementary DNA strands.

Thermal error removal
We use precise control of synthesis site temperature to melt and remove error-containing sequences, ensuring accuracy of the final DNA.
